Author
Author  Correspondence author
Correspondence author
Maize Genomics and Genetics, 2024, Vol. 15, No. 5 doi: 10.5376/mgg.2024.15.0023
Received: 03 Aug., 2024 Accepted: 17 Sep., 2024 Published: 08 Oct., 2024
Zhou J., and Xu M.L., 2024, Comparative genomics of maize: insights into evolution and function, Maize Genomics and Genetics, 15(5): 239-246 (doi: 10.5376/mgg.2024.15.0023)
Comparative genomics of maize (Zea mays) has provided transformative insights into the evolutionary processes and functional mechanisms that have shaped this important crop. Through the analysis of whole-genome sequences, gene family expansions, and structural variations, researchers have deepened their understanding of maize's domestication, adaptation, and diversification. Key findings include the identification of conserved regulatory elements that play critical roles in gene expression, the impact of gene duplication events that have led to functional diversification, and the discovery of genomic regions linked to traits such as yield, stress tolerance, and disease resistance. These studies have also revealed the evolutionary relationships between maize and other members of the grass family (Poaceae), contributing to a broader understanding of plant evolution. The application of these findings extends to maize breeding and biotechnology, where genomic data is being used to develop more resilient and higher-yielding maize varieties. As genomic technologies such as CRISPR, pan-genomics, and multi-omics integration continue to advance, the field of maize comparative genomics is poised to play a critical role in addressing future challenges in agriculture and food security.
1 Introduction
Maize (Zea mays), one of the most significant crops globally, plays a dual role as both a staple food source and a model organism for plant genetics. Its economic importance is underscored by its use in food, feed, and biofuel production, making it essential for agricultural sustainability. Beyond agriculture, maize has been a powerful tool in genomics research due to its genetic diversity and complex genome structure. The domestication of maize began approximately 9 000 years ago in the Balsas River region of southern Mexico, where it was derived from its wild ancestor teosinte. Over millennia, selective breeding and adaptation to diverse environments have enhanced its genetic complexity, making it an excellent model for studying genome evolution and gene function in plants (Hufford et al., 2021).
Comparative genomics is a crucial approach in plant biology that examines the similarities and differences between the genomes of different species. This method helps identify conserved and divergent genomic regions, enabling researchers to uncover evolutionary processes and functional elements that define species-specific traits (Huang and Hong, 2024). In maize, comparative genomics has revealed significant insights into genome architecture, such as gene duplications, deletions, and chromatin rearrangements, which contribute to the plant's phenotypic diversity and adaptability. Studies comparing maize to closely related species, like sorghum and rice, have highlighted important evolutionary trends, including genome fractionation and recombination, that have shaped maize’s genomic landscape. These findings are essential for understanding maize's unique evolutionary history and its potential for agricultural improvement (Lozano et al., 2021; Tian et al., 2021).
This study utilizes comparative genomics to deepen the understanding of maize evolution and functional genomics. By analyzing structural variations, gene expression, and methylation patterns across different maize lines, it aims to identify genetic elements critical for traits such as stress tolerance and pathogen resistance. The research also provides new insights into maize adaptability, offering guidance for strategies to enhance crop resilience and productivity in response to environmental changes. This study will not only contribute to fundamental knowledge in plant biology but also offer practical applications for improving maize cultivation and breeding practices.
2 Methods for Comparative Maize Genomics
2.1 Genomic data collection and sources
In comparative maize genomics, various types of genomic data are utilized, including whole-genome sequences, transcriptomes, and methylomes. The reference genome of maize has been continuously improved with new technologies such as Single Molecule Real-Time (SMRT) sequencing, which enables better assembly of repetitive regions and centromeres. The maize B73 genome, for example, has seen improvements in both contig length and annotation quality, with over 130 000 transposable elements identified, making it a key resource for comparative genomic studies (Jiao et al., 2017). Additionally, the Maize Genetics and Genomics Database (MaizeGDB) provides extensive resources for maize genomes, including data from multiple inbred lines and tools for genome comparison and analysis (Portwood et al., 2018).
2.2 Analytical techniques in comparative genomics
Various bioinformatics tools and techniques are applied to compare maize genomes. Sequence alignment tools, such as GSAlign, are essential for efficient and accurate comparisons across multiple genomes (Lin and Hsu, 2019). Synteny analysis, which involves identifying conserved gene order across species, plays a crucial role in detecting orthologs and paralogs, especially with tools like SynFind. This web-based tool allows for the identification of syntenic regions, providing critical insights into gene evolution and genomic conservation across different maize genomes (Tang et al., 2015). These methods enable researchers to determine orthologous genes and track gene loss or transposition events, offering deep insights into maize genome evolution.
2.3 Addressing challenges in comparative genomics
Maize genomics presents challenges due to the complexity of its genome, including polyploidy and extensive repetitive elements. Polyploidy has resulted in multiple gene duplications, making it difficult to distinguish between paralogs and orthologs. Tools like SynerClust address this by incorporating synteny information to improve the accuracy of ortholog identification while managing large-scale datasets (Georgescu et al., 2018). Additionally, advances in sequencing technologies, such as SMRT sequencing, have improved the resolution of repetitive regions, helping researchers assemble more accurate maize genomes and tackle challenges associated with genome complexity (Jiao et al., 2017).
3 Evolutionary Insights from Maize Comparative Genomics
3.1 Phylogenetic relationships within the grasses
Comparative genomics has refined our understanding of the phylogeny within the grass family (Poaceae). By aligning genome sequences of key grasses, researchers have uncovered variable evolutionary rates among Poaceae species, with slower substitution rates in rice and significantly faster rates in other grasses. This genomic evidence has altered the estimated dates for key evolutionary events, such as the divergence between maize and sorghum, which coincided with polyploidization about 96 million years ago, suggesting that polyploidization may have directly contributed to grass speciation (Wang et al., 2015).
3.2 Gene family evolution
Gene family expansions and contractions have played a significant role in maize evolution. Studies show that gene duplication through whole-genome duplication (WGD) and subsequent subfunctionalization have driven the diversification of gene families in maize. For example, remorin genes, which are involved in stress responses, have expanded in the Poaceae family due to WGD and segmental duplications, highlighting the evolutionary adaptation of maize to environmental stresses (Wang et al., 2022). Other studies have identified specific expansions in genes related to flowering time and stress response, showing that gene duplications contribute to maize's adaptation to diverse environments (Meng and Yang, 2019).
3.3 Insights into domestication and adaptation
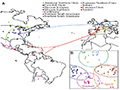
Comparative genomics has shed light on the domestication and adaptation of maize. Genomic analyses have identified domestication genes such as tb1 and zcn8, which control important traits like reduced tillering and flowering time. Additionally, adaptation to different environments has been facilitated by selection on genes related to stress responses (Huang, 2024). For instance, studies show that maize landraces adapted to high altitudes possess genetic changes that confer advantages such as earlier flowering times, driven by selection in both Mesoamerican and South American highlands (Takuno et al., 2015). Moreover, genomic segments displaying high heterozygosity in European maize are linked to adaptation traits such as cold tolerance and flowering time (Figure 1) (Brandenburg et al., 2017).
|
Figure 1 Sample locations, genetic structuring and inferred routes of maize migration (Adopted from Brandenburg et al., 2017) Image caption: A: Geographic location of 66 landraces from which lines originated with colors of dots designating genetic groups defined a priori-a Spanish line for which the geographical coordinates are unknown is not represented. Arrows indicate inferred routes of maize migration with admixed groups displaying two arrows. Colors of the arrows correspond to the recipient group; B: Principal Component Analysis computed on 500k non-genic SNPs. Corresponding ellipses indicate the 95% CI of the Mahalanobis distance (Adopted from Brandenburg et al., 2017) |
4 Functional Genomics in Maize
4.1 Identification of functional elements
Comparative genomics has been instrumental in identifying conserved regulatory elements in maize. These elements, such as conserved non-coding sequences (CNSs), play critical roles in gene regulation, including chromatin interaction sites and cis-regulatory elements. In the Andropogoneae tribe, which includes maize, CNSs have been associated with DNA replication, methylation, and histone modification. Studies show that variations in CNSs can lead to significant differences in gene expression, with the absence of CNSs contributing to the loss of gene expression in certain contexts (Song et al., 2020). Another key discovery is that long-range cis-regulatory elements are widespread in the maize genome and contribute to the regulation of genes controlling agronomic traits. These elements, located far from their target genes, are involved in complex chromatin loops that influence gene expression (Ricci et al., 2019).
4.2 Gene expression and regulation
Gene expression regulation in maize varies significantly between tissues and developmental stages. Comparative analyses using RNA-Seq data have revealed how regulatory networks, including transcription factor binding motifs and non-coding RNAs, contribute to organ-specific gene expression. Open chromatin assays have identified chromatin-accessible regions associated with regulatory elements that control gene expression during maize inflorescence development. These elements shape the development of male and female reproductive organs by guiding tissue-specific transcription factors (Parvathaneni et al., 2020). The integration of transcriptome data has enabled the construction of gene regulatory networks (GRNs), which further explain how gene expression is controlled in various maize tissues such as leaves, roots, and seeds (Huang et al., 2018).
4.3 Functional diversification of genes
Gene duplication events have been a major driver of functional diversification in maize. Following whole-genome duplication (WGD) events, duplicated genes in maize have evolved different functions, especially through subfunctionalization in different tissues. Studies on maize subgenomes indicate that some duplicated genes experience stronger purifying selection, leading to divergence in gene expression and functional roles. This subgenomic evolution contributes to maize’s ability to adapt to diverse environments (Pophaly and Tellier, 2015). A notable case of functional diversification involves the α-zein gene family, where tandem duplications have resulted in variation in gene expression across different maize haplotypes, highlighting both conservation and diversification in the regulatory patterns of duplicated genes (Hurst et al., 2021).
5 Applications of Comparative Genomics in Maize Improvement
5.1 Breeding for disease resistance
Comparative genomics has been extensively used to identify genes associated with disease resistance in maize, including resistance to Fusarium ear rot, maize lethal necrosis (MLN), and fall armyworm (FAW). Genome-wide association studies (GWAS) and quantitative trait locus (QTL) mapping have identified several genomic loci linked to disease resistance. For example, multiple SNP markers have been associated with resistance to MLN, gray leaf spot, and turcicum leaf blight in different environments, providing valuable insights for marker-assisted breeding programs (Sadessa et al., 2022). Similarly, resistance to Fusarium ear rot has been linked to specific SNPs located on maize chromosomes 2, 3, 4, 5, 9, and 10, offering insights into the development of resistant maize lines (Chen et al., 2016).
5.2 Enhancing stress tolerance
Abiotic stress tolerance in maize, particularly drought and heat tolerance, has been a significant focus of comparative genomics studies. Quantitative trait loci (QTL) mapping and genome-wide association studies (GWAS) have identified regions of the genome linked to drought stress tolerance. Recent advancements in molecular breeding techniques, including marker-assisted selection (MAS) and genomic selection, have been used to develop drought-tolerant maize varieties, particularly in regions like Sub-Saharan Africa where drought is a significant challenge (Wang et al., 2019). Moreover, genetic tools such as CRISPR-Cas9 and transgenic approaches have contributed to the development of maize lines with enhanced tolerance to multiple stresses (Figure 2) (Malenica et al., 2021).
|
Figure 2 Most important stress factors in maize (Adopted from Malenica et al., 2021) |
5.3 Improving nutritional content
Comparative genomic approaches have also been applied to enhance the nutritional content of maize. Research has identified key genes linked to important nutritional traits such as protein and vitamin content. For example, functional genes associated with kernel-related traits have been mapped onto specific chromosomes using marker-assisted selection. These genes are being exploited to improve the nutritional value of maize kernels, including improving the content of quality proteins and micronutrients. Metabolite quantitative trait loci (mQTL) analyses have also revealed important loci associated with higher nutritional content, including those related to metabolites that improve grain quality (Li et al., 2019).
6 Future Directions in Maize Comparative Genomics
6.1 Integration of multi-omics data
The integration of multi-omics data-combining genomic, transcriptomic, proteomic, and metabolomic data-offers a comprehensive view of maize biology, helping to link genes to complex traits. These approaches are critical for understanding growth, yield, and responses to environmental stress in maize. Multi-omics platforms, including transcriptomics and proteomics, have been successfully implemented to uncover molecular mechanisms driving phenotypic variation in crops like maize. One challenge in multi-omics integration is the complexity of analyzing large datasets from multiple biological layers, which requires advanced computational methods and data fusion techniques (Yang et al., 2021). Systems biology tools, like pathway analysis and network-based integration, can provide more accurate models for crop improvement by incorporating data from multiple omics layers (Paczkowska et al., 2018).
6.2 Advances in genomic technologies
Emerging genomic technologies such as CRISPR, long-read sequencing, and pan-genomics are revolutionizing maize research. Long-read sequencing, exemplified by technologies like PacBio and Oxford Nanopore, is essential for resolving highly repetitive regions in the maize genome and constructing gapless telomere-to-telomere assemblies. These advancements provide more complete and accurate genome assemblies that enhance comparative genomic studies (Liu et al., 2020). CRISPR-Cas9 has emerged as a key tool for genome editing in maize, offering precise manipulation of genes responsible for yield, stress tolerance, and disease resistance. Moreover, pan-genomics-the study of the collective genome of a species-enables researchers to capture the full genetic diversity of maize, uncovering rare and novel alleles that can be utilized in breeding programs (Zhang et al., 2022).
6.3 Addressing unresolved questions
Despite advancements in maize genomics, several questions remain unresolved. For instance, the evolutionary processes that underlie the domestication and adaptation of maize to diverse environments require further exploration. Uncovering the functional roles of non-coding regions and their contributions to trait expression and adaptation is an ongoing challenge. Additionally, understanding how maize genomes interact with their environments, particularly under climate change, is crucial for improving resilience and yield. Comparative genomics, coupled with multi-omics approaches, can help fill these gaps by providing deeper insights into gene-environment interactions and adaptive evolution (Westhues et al., 2017).
7 Concluding Remarks
Comparative genomics in maize has provided numerous insights that have reshaped our understanding of the evolutionary relationships, gene function, and adaptive traits in this crop. Studies have identified key gene families involved in disease resistance, stress tolerance, and nutrient uptake, leveraging genomic data for breeding applications. The identification of conserved regulatory elements, gene duplication events, and structural genomic variations have furthered our comprehension of maize biology. These findings are not only critical for evolutionary biology but also hold immense value for agricultural applications. For example, understanding the genomic underpinnings of traits like drought tolerance and yield has already enabled more targeted breeding efforts, as shown in multiple genome-wide association studies and QTL analyses.
The insights gained from comparative genomics are being directly applied to maize breeding and biotechnology. CRISPR and other genome-editing technologies are now commonly used to introduce targeted improvements in traits such as disease resistance, stress tolerance, and yield. By leveraging comparative genomics, breeders can better identify critical genes that contribute to these traits, and genomic prediction models are improving the efficiency of hybrid maize breeding programs. Furthermore, innovations in pan-genomics have highlighted the diversity within maize populations, revealing rare alleles that may have gone unnoticed in traditional breeding, thus expanding the toolbox for developing maize varieties better suited to specific environments.
Continued research in maize comparative genomics will be crucial for addressing unresolved questions related to the plant's adaptation, domestication, and genetic diversity. Emerging technologies like long-read sequencing and single-molecule real-time sequencing are expected to further enhance our understanding of complex genomic regions, including repetitive elements and centromeres. The broader impact of maize genomics extends beyond maize, offering a model for other crops where similar approaches can be applied. Future research must focus on integrating multi-omics data to fully capture the complexity of maize biology and translate these findings into tangible improvements in crop performance under various environmental conditions.
Acknowledgments
The publisher sincerely thanks the anonymous peer reviewers for their thorough evaluation of this manuscript.
Conflict of Interest Disclosure
The authors affirm that this research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
Brandenburg J.T., Mary-Huard T., Rigaill G., Hearne S., Corti H., Joets J., Vitte C., Charcosset A., Nicolas S.D., and Tenaillon M.I,, 2017, Independent introductions and admixtures have contributed to adaptation of European maize and its American counterparts, PLoS Genetics, 13(6): e1006666.
https://doi.org/10.1371/journal.pgen.1006666
PMID: 28301472 PMCID: PMC5373671
Chen J.F., Shrestha R., Ding J.Q., Zheng H.J., Mu C.H., Wu J.Y., and Mahuku G., 2016, Genome-wide association study and QTL mapping reveal genomic loci associated with Fusarium ear rot resistance in tropical maize germplasm, G3: Genes|Genomes|Genetics, 6(12): 3803-3815.
https://doi.org/10.1534/g3.116.034561
PMID: 27742723 PMCID: PMC5144952
Georgescu C.H., Manson A.L., Griggs A.D., Desjardins C.A., Pironti A., Wapinski I., Abeel T., Haas B.J., and Earl A.M., 2018, SynerClust: a highly scalable, synteny-aware orthologue clustering tool, Microbial Genomics, 4(11): e000231.
https://doi.org/10.1099/mgen.0.000231
PMID: 30418868 PMCID: PMC6321874
Huang D.D., 2024, CRISPR/Cas9 genome editing in legumes: opportunities for functional genomics and breeding, Legume Genomics and Genetics, 15(4): 199-209.
https://doi.org/10.5376/lgg.2024.15.0020
Huang J., Zheng J.F., Yuan H., and McGinnis K., 2018, Distinct tissue-specific transcriptional regulation revealed by gene regulatory networks in maize, BMC Plant Biology, 18(1): 111.
https://doi.org/10.1186/s12870-018-1329-y
PMID: 29879919 PMCID: PMC6040155
Huang W.Z., and Hong Z.M., 2024, Marker-assisted selection in cassava: from theory to practice, Plant Gene and Trait, 15(1): 33-43.
https://doi.org/10.5376/pgt.2024.15.0005
Hurst P., Schnable J.C., and Holding D.R., 2021, Tandem duplicate expression patterns are conserved between maize haplotypes of the α‐zein gene family, Plant Direct, 5(9): e346.
https://doi.org/10.1002/pld3.346
Hufford M.B., Seetharam A.S., Woodhouse M.R., Chougule K., Ou S., Liu J., Ricci W.A., Guo T., Olson A.J., Qiu Y., Coletta R.D. Tittes S.B., Hudson A.I., Marand A.P., Wei S., Lu Z., Wang B., Tello-Ruiz M., Piri R.D., Wang N., Kim D.W., Zeng Y., O’Connor C.H., Li X., Gilbert A. M., Baggs E., Krasileva K., Portwood J.L., Cannon E.K.S., Andorf C.M., Manchanda N., Snodgrass S., Hufnagel D.E., Jiang Q., Pedersen S., Syring M.L., Kudrna D., Llaca V., Fengler K., Schmitz R., Ross-Ibarra J., Yu J., Gent J., Hirsch C., Ware D., and Dawe R.K., 2021, De novo assembly, annotation, and comparative analysis of 26 diverse maize genomes, Science, 373: 655-662.
https://doi.org/10.1126/science.abg5289
Jiao Y.P., Peluso P., Shi J.H., Liang T., Stitzer M.C., Wang B., Campbell M.S., Stein J.C., Wei X.H., Chin C.S., Guill K., Regulski M., Kumari S., Olson A., Gent J., Schneider K.L., Wolfgruber T.K., May M.R., Springer N.M., Antoniou E., McCombie W.R., Presting G.G., McMullen M., Ross-Ibarra J., Dawe R.K., Hastie A., Rank D.R., and Ware D., 2017, Improved maize reference genome with single-molecule technologies, Nature, 546(7659): 524-527.
https://doi.org/10.1038/nature22971
PMID: 28605751 PMCID: PMC7052699
Li K., Wen W.W., Alseekh S., Yang X.H., Guo H., Li W.W., Wang L.X., Pan Q.C., Zhan W., Liu J., Li Y.H., Wu X., Brotman Y., Willmitzer L., Li J.S., Fernie A.R., and Yan J.B., 2019, Large-scale metabolite quantitative trait locus analysis provides new insights for high-quality maize improvement, The Plant Journal, 99(2): 216-230.
https://doi.org/10.1111/tpj.14317
PMID: 30888713
Lin H.N., and Hsu W., 2019, GSAlign: an efficient sequence alignment tool for intra-species genomes, BMC Genomics, 21(1): 182.
https://doi.org/10.1186/s12864-020-6497-4
PMID: 32093618 PMCID: PMC7041101
Liu C.H. Song W., Luo Y.F., Gao S.H., Zhang R.Y,, Shi Z., Wang X.Q., Wang R.H., Wang F.G., Wang J.D., Zhao Y.X., Su A.G., Wang S., Zhang Y.X., Ge J.R., Yuan Y., Bi X.C., Yan J.B., Wang Y.D., Hu S.N., and Zhao J.T., 2019, The Huangzaosi maize genome provides insights into genomic variation and improvement history of maize, Molecular Plant, 12(3): 402-409.
https://doi.org/10.1016/j.molp.2019.02.009
PMID: 30807824
Liu J.N., Seetharam A.S., Chougule K., Ou S.J., Swentowsky K.W., Gent J.I., Llaca V., Woodhouse M.R., Manchanda N., Presting G.G., Kudrna D.A., Alabady M., Hirsch C.N., Fengler K.A., Ware D., Michael T.P., Matthew B Hufford M.B., and Dawe R.K., 2020, Gapless assembly of maize chromosomes using long-read technologies, Genome Biology, 21(1): 121.
https://doi.org/10.1186/s13059-020-02029-9
PMID: 32434565 PMCID: PMC7238635
Lozano R., Gazave E., dos Santos J.P.D., Stetter M.G., Valluru R., Bandillo N., Fernandes S.B., Brown P.J., Shakoor N., Mockler T.C., Cooper E.A., Buckler E.S., Ross-Ibarra J., Buckler E., and Gore M.A., 2021, Comparative evolutionary genetics of deleterious load in sorghum and maize, Nature Plants, 7(1): 17-24.
https://doi.org/10.1038/s41477-020-00834-5
PMID: 33452486
Malenica N., Dunić J.A., Vukadinović L., Cesar V., and Šimić D., 2021, Genetic approaches to enhance multiple stress tolerance in maize, Genes, 12(11): 1760.
https://doi.org/10.3390/genes12111760
PMID: 34828366 PMCID: PMC8617808
Meng Y., and Yang R.L., 2019, Comparative analysis of gene family size provides insight into the adaptive evolution of vertebrates, Yi Chuan, 41(2): 158-174.
https://doi.org/10.16288/j.yczz.18-182
PMID: 30803946
Paczkowska M., Barenboim J., Sintupisut N., Fox N.S., Zhu H.L., Abd-Rabbo D., Mee M.W., Boutros P.C., PCAWG Drivers and Functional Interpretation Working Group, Jüri Reimand J., and PCAWG Consortium, 2018, Integrative pathway enrichment analysis of multivariate omics data, Nature Communications, 11(1): 735.
https://doi.org/10.1038/s41467-019-13983-9
PMID: 32024846 PMCID: PMC7002665
Parvathaneni R.K., Bertolini E., Shamimuzzaman M., Vera D.L., Lung P.Y., Rice B.R., Zhang J.F., Brown P.J., Lipka A.E., Bass H.W., and Eveland A.L., 2020, The regulatory landscape of early maize inflorescence development, Genome Biology, 21(1): 165.
https://doi.org/10.1186/s13059-020-02070-8
PMID: 32631399 PMCID: PMC7336428
Pophaly S.D., and Tellier A., 2015, Population level purifying selection and gene expression shape subgenome evolution in maize, Molecular Biology and Evolution, 32(12): 3226-3235.
https://doi.org/10.1093/molbev/msv191
PMID: 26374232
Portwood J.L., Woodhouse M.R., Cannon E.K.S., Gardiner J.M., Harper L.C., Schaeffer M.L., Walsh J.R., Sen T.Z., Cho K.T., Schott D.A., Braun B.L., Dietze M., Dunfee B., Elsik C.G., Manchanda N., Coe E., Sachs M., Stinard P., Tolbert J., Zimmerman S., and Andorf C.M., 2018, MaizeGDB 2018: The maize multi-genome genetics and genomics database, Nucleic Acids Research, 47: D1146-D1154.
https://doi.org/10.1093/nar/gky1046
PMID: 30407532 PMCID: PMC6323944
Ricci W.A., Lu Z.F., Ji L., Marand A.P., Ethridge C.L., Murphy N.G., Noshay J.M., Galli M., Mejía-Guerra M.K., Colomé-Tatché M., Johannes F., Rowley M.J., Corces V.G., Zhai J.X., Scanlon M.J., Buckler E.S., Gallavotti A., Springer N.M., Schmitz R.J., and Zhang X.Y., 2019, Widespread long-range cis-regulatory elements in the maize genome, Nature Plants, 5(12): 1237-1249.
https://doi.org/10.1038/s41477-019-0547-0
PMID: 31740773 PMCID: PMC6904520
Sadessa K., Beyene Y., Ifie B., Suresh L., Olsen M.S., Ogugo V., Wegary D., Tongoona P., Danquah E., Offei S.K., Prasanna B.M., and Gowda M., 2022, Identification of genomic regions associated with agronomic and disease resistance traits in a large set of multiple DH populations, Genes, 13(2): 351.
https://doi.org/10.3390/genes13020351
PMID: 35205395 PMCID: PMC8872035
Song B.X., Wang H., Wu Y.Y., Rees E., Gates D.J,, Burch M., Bradbury P.J., Ross-Ibarra J., Kellogg E.A., Hufford M.B., Romay M.C., and Buckler E., 2020, Constrained non-coding sequence provides insights into regulatory elements and loss of gene expression in maize, bioRxiv, 12: 2020-07.
https://doi.org/10.1101/2020.07.11.192575
Takuno S., Ralph P., Swarts K., Elshire R.J., Glaubitz J.C., Buckler E.S., Hufford M.B., and Ross-Ibarra J., 2015, Independent molecular basis of convergent highland adaptation in maize, Genetics, 200(4): 1297-1312.
https://doi.org/10.1534/genetics.115.178327
PMID: 26078279 PMCID: PMC4571994
Tang H.B., Bomhoff M.D., Briones E., Zhang L.S., Schnable J.C., and Lyons E., 2015, SynFind: compiling syntenic regions across any set of genomes on demand, Genome Biology and Evolution, 7(12): 3286-3298.
https://doi.org/10.1093/gbe/evv219
PMID: 26560340 PMCID: PMC4700967
Tian L., Ku L.X., Yuan Z., Wang C.L., Su H.H., Wang S.X., Song X.C., Dou D.D., Ren Z.Z., Lai J.S., Liu T., Du C.G., and Chen Y.H., 2021, Large-scale reconstruction of the chromatin structures of maize temperate and tropical inbred lines, Journal of Experimental Botany, 72(10): 3582-3596.
https://doi.org/10.1093/jxb/erab087
PMID: 33677565
Wang N., Liu B.J., Liang X.L., Zhou Y.H., Song J., Yang J., Yong H.J., Weng J.F., Degui Zhang D.G., Li M.S., Nair S., Vicente F.S., Hao Z.F., Xuecai Zhang X.C., and Li X.H., 2019, Genome-wide association study and genomic prediction analyses of drought stress tolerance in China in a collection of off-PVP maize inbred lines, Molecular Breeding, 39: 1-16.
https://doi.org/10.1007/s11032-019-1013-4
Wang X.Y., Wang J.P., Jin D.C., Guo H., Lee T.H., Liu T., and Paterson A.H., 2015, Genome alignment spanning major poaceae lineages reveals heterogeneous evolutionary rates and alters inferred dates for key evolutionary events, Molecular Plant, 8(6): 885-898.
https://doi.org/10.1016/j.molp.2015.04.004
PMID: 25896453
Wang Y.Q., Li J.Q., Li M.Y., Li Y.T., Zhao Z.B., Li C., and Yue J., 2022, Genome-wide characterization of remorin genes in foxtail millet reveals their evolution and response to abiotic stresses, Diversity, 14(9): 711.
https://doi.org/10.3390/d14090711
Westhues M., Schrag T.A., Heuer C., Thaller G., Utz H.F., Schipprack W., Thiemann A., Seifert F., Ehret A., Schlereth A., Stitt M., Nikoloski Z.,Willmitzer L., Schön C.C., Scholten S., and Melchinger A.E., 2017, Omics-based hybrid prediction in maize, Theoretical and Applied Genetics, 130: 1927-1939.
https://doi.org/10.1007/s00122-017-2934-0
PMID: 28647896
Yang Y.D., Saand M.A., Huang L.Y., Abdelaal W.B., Zhang J., Wu Y., Li J., Sirohi M.H., and Wang F.Y., 2021, Applications of multi-omics technologies for crop improvement, Frontiers in Plant Science, 12: 563953.
https://doi.org/10.3389/fpls.2021.563953
PMID: 34539683 PMCID: PMC8446515
Zhang F., Xue H.Z., Dong X.R., Li M., Zheng X.M., Li Z.K., Xu J.L., Wang W.S., and Wei C.C., 2022, Long-read sequencing of 111 rice genomes reveals significantly larger pan-genomes, Genome Research, 32: 853-863.
https://doi.org/10.1101/gr.276015.121
PMID: 35396275 PMCID: PMC9104699

. PDF(514KB)
. FPDF(win)
. FPDF(mac)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Jin Zhou
. Minli Xu
Related articles
. Comparative genomics
. Maize evolution
. Gene duplication
. Domestication
. Maize breeding
Tools
. Email to a friend
. Post a comment